3.3: Molecular toolbox – Neural Circuits (The Basics)
- Page ID
- 10651
- Precision targeting of neurons
- Optogenetics the basics
- Channelrhodopsins
- Inhibitory Opsins
Precision Targeting of Neurons
Neurons don’t work in isolation and they typically make synaptic connections that resemble circuits. Most investigations of behavior and development have relied on mapping the neural circuits within the brain and CNS. Understanding the connectivity of these circuits allow neuroscientists to understand behavior and pathologies (Figure \(\PageIndex{1}\).). In the past, this has meant lesioning or destroying parts of these circuits, or electrophysiologically stimulating these circuits and examining the resulting behavior. These older methods have allowed us to dissect the detailed workings of the neural circuits underlying natural behavior, and have also enabled us to understand how some neural circuits become dysfunctional in disease states such as Parkinson’s disease or epilepsy.
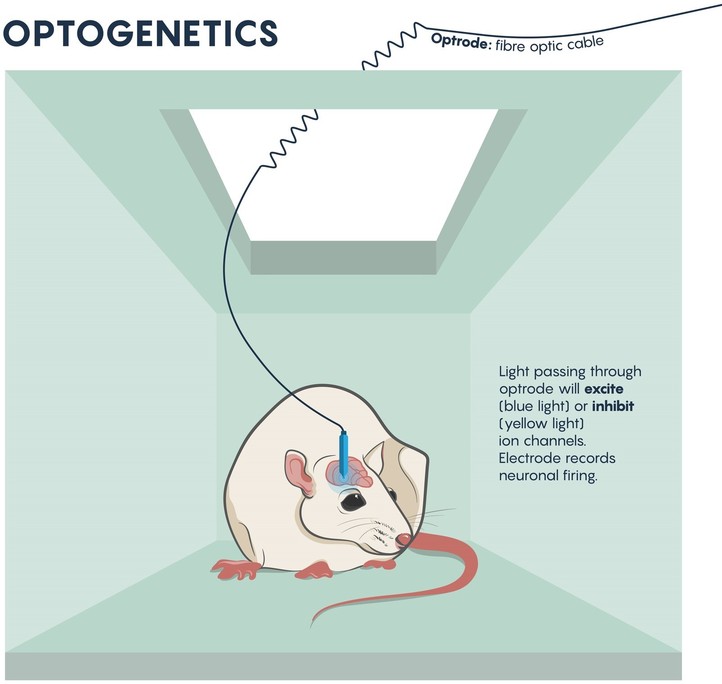
Figure \(\PageIndex{1}\): The beginnings of brain circuit studies in mice using optrodes to excite or inhibit ion channels. Adapted from Nature 2016 (open access cc by 4).
The vertebrate brain (mice, rats, primates and humans), contains many different cell types with distinct molecular expression patterns, physiological activity, and topological connectivity, which are intermingled in a highly heterogeneous network. Studying specific groups of neurons in this milieu becomes very challenging and scientists using lesion, stimulation and tracing studies were never sure about the spatial (i.e. were only specific neurons affected) and temporal (i.e. can the lesion be reversed, allowing for reversal of behaviour).
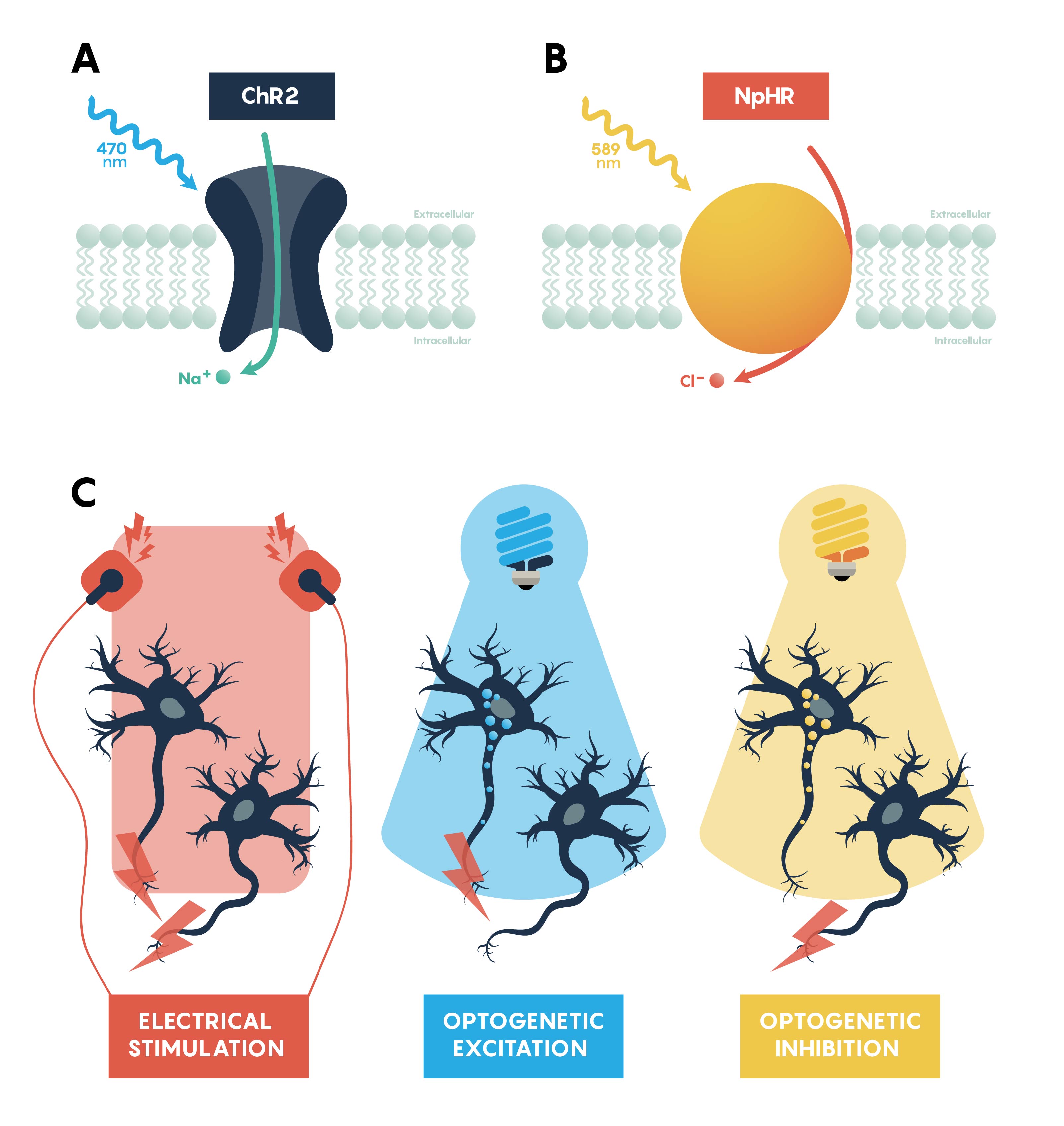
Figure \(\PageIndex{2}\). Panels summarizing the features of excitatory, ChR2, (A) and inhibitory, NpHR, (B) opsins. Panel C showing that all neurons would be excitable using a standard electrode. However, optogenetic excitation (blue light) and inhibition (yellow light) only work on specific neurons that express the appropriate opsin.
The questions around specificity and temporal reversibility changed with the introduction of optogenetics. Since the late 2000s, optogenetics has ushered in a new era of potent and targeted control over multiple aspects of neural function. Genetic and optical methods applied together allow tight spatial and temporal control of the activity of specific kinds of neurons in the living brain, a revolutionary advance that will allow us to achieve an unprecedented understanding of neural circuit function in behaving animals. Using this technique, neurons are first genetically engineered (using a variety of mechanisms, described later) to express light-sensitive proteins (opsins). When these neurons are then illuminated with light of the correct frequency they will be transiently activated or inhibited or their signaling pathways will be modulated, depending on the particular kind of opsin that was chosen for expression. Cell type–specific expression is typically achieved with transgenic animals, viral vectors, or a combination, and spatially restricted light application allows for further refinement in targeting to specific brain regions. Light can be applied in a variety of temporal patterns in order to optimally influence neuronal function (permitting experimental control of spike frequency and burstiness, among other parameters), and may be restricted to specific short behavioral periods of examination.
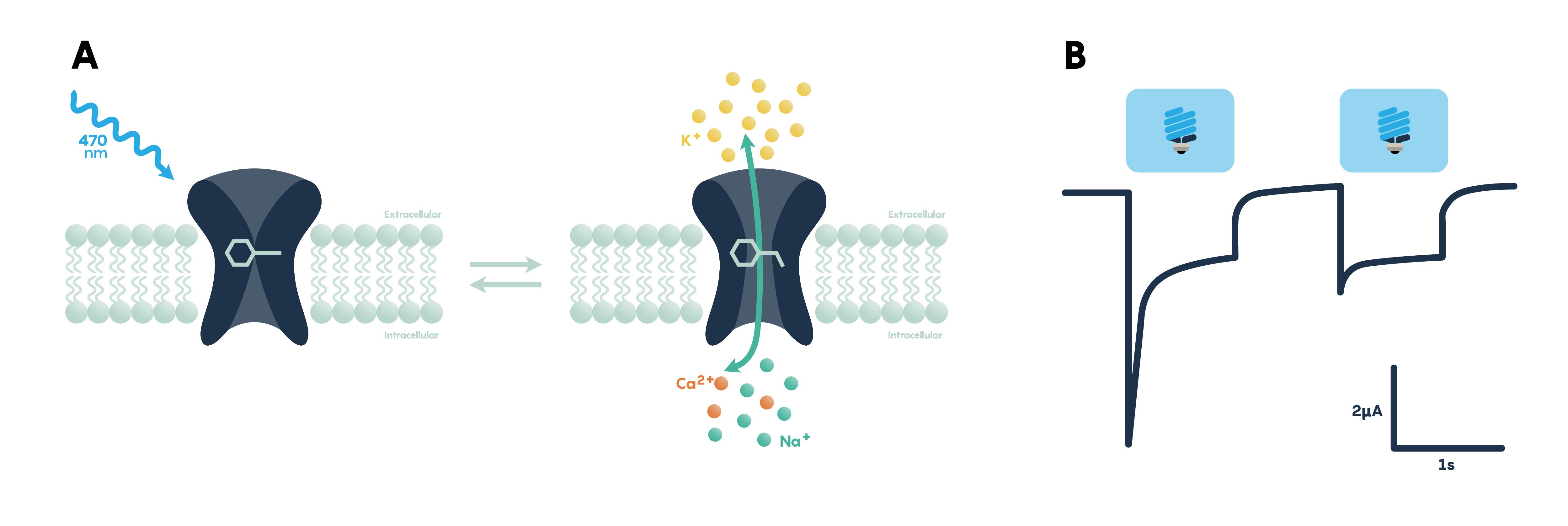
Figure \(\PageIndex{3}\). Panel A showing the excitatory effect of blue light on a membrane-bound opsin functioning as a channel. Panel B showing action potentials in response to blue (excitatory) light.
Optogenics: The Basics
What Are Optogenetic Actuators?
Optogenetic actuators are proteins that modify the activity of the cell in which they are expressed when that cell is exposed to light (Figure \(\PageIndex{3}\).). These actuators can be used to induce single or multiple action potentials (which can be organized into regular spike trains or which can be pseudo-random at a user-controlled rate), suppress neural activity, or modify biochemical signaling pathways, with millisecond control over the timing of events. The most powerful and widely used actuators are opsins—naturally occurring light-sensitive transmembrane proteins—that are found in a variety of organisms ranging from microbes to primates, and that can be used as found in nature or engineered to optimize functioning. Naturally occurring opsins can be broadly categorized into two major classes: microbial opsins (Type I) and vertebrate opsins (Type II). Type I opsins are found in prokaryotic and eukaryotic microbial organisms, including bacteria, archaea, and algae, and are composed of a single membrane–bound protein component that functions as a pump or channel. These opsins are used by their host microorganisms for a variety of functions, including navigation towards sources of energy and away from hazardous environments, and control the intracellular concentrations of a variety of ions and the beating of flagella.
Type I opsins were used in the first optogenetics experiments to control neuronal function, both because of the ease of genetic engineering using a single component protein and because of their faster kinetics, and remain the primary (but not exclusive) source for new natural and engineered opsins.

Figure \(\PageIndex{4}\). Opsins are membrane-bound proteins that are activated with light, which results in cell activation (depolarization), inhibition (hyperpolarization), or modulation of intracellular signaling cascades (panel A). Illustrated here are ChR2 (a cation channel used to stimulate neural activity), iC1C2 (a newly developed chloride channel used to inhibit neural activity), eNpHR3.0 (a chloride pump used to inhibit neural activity), eBR (a proton pump used to inhibit neural activity), and OptoXR (a G protein–coupled receptor used to modulate intracellular signaling cascades). (Panel B) Cell-attached and whole-cell recordings from a neuron expressing both ChR2 and NpHR. Note that individual spikes can be elicited with a short pulse of blue light (which activates ChR2) and that these spikes can be blocked with continuous yellow light (which activates NpHR). Panel A adapted with permission from Fenno et al., 2011, and panel B adapted with permission from Zhang et al., 2007.
How Do Optogenetic Actuators Work?
Opsins of both types require retinal, a form of vitamin A that isomerizes upon absorption of a photon, in order to function. When retinal binds to the opsin the retinal-opsin complex becomes light sensitive, and if a photon strikes the retinal in this state its resulting photoisomerization will induce a conformational change in the opsin. This leads to channel opening or pump activation, a change in membrane potential, and ultimately the activation or inhibition of neuronal activity. Therefore, retinal must be present in order for optogenetic actuators to function. Fortunately, particularly for the early proof-of-principle experiments, retinal is already present in sufficient quantities in mammalian neural tissue to permit the use of optogenetic tools without exogenous retinal supplementation. However, invertebrate model systems such as Drosophila do need retinal supplementation through their diet in order for optogenetic effectors to function. Here we review the different classes of optogenetic actuators, grouped by their effect on neural activity or signaling.
Optogenetic Stimulation of Neural Activity: How to Turn Neurons “On”
Channelrhodopsins
Channelrhodopsins (ChRs) are light-gated ion channels discovered in Chlamydomonas reinhardtii, a unicellular green alga. The first use of a microbial opsin to control the spiking activity of neurons utilized Channelrhodopsin-2 (ChR2), one of two channelrhodopsins expressed by this organism. ChR2 is a light-gated nonspecific cation channel which, when illuminated with blue light, opens and allows the passage of cations and the subsequent depolarization of the cell. In 2005 ChR2 was introduced into cultured hippocampal neurons and successfully used to control spiking activity with fine temporal precision. As demonstrated by this pioneering paper, very brief (millisecond) pulses of blue light can be used to induce single action potentials in ChR2-expressing neurons, and spiking activity driven by the activation of this opsin can be controlled with high precision at frequencies approaching 30 spikes per second.
Optogenetic Inhibition of Neuronal Activity: How to Turn Neurons “Off”
Chloride Pumps
Inhibiting neuronal activity in neural circuits can complement excitatory tools by allowing investigators to test the role of individual neuronal circuit components. One of the most efficient and widely used optogenetic inhibitory opsins, NpHR, is a halorhodopsin from the archaeon Natronomonas pharaonis. NpHR pumps chloride ions into the cell upon long wavelength light activation, resulting in hyperpolarization. Genetic engineering has led to a series of revisions producing eNpHR3.0, an opsin with improved surface membrane localization and a larger photocurrent. With an excitation maximum at 590nm, eNpHR3.0 can be driven by green, yellow, or red wavelengths of light, enabling the use of less expensive laser systems.
Proton Pumps
Proton pumps can also be used to inhibit neurons through hyperpolarization, by pumping protons out of the cell, and have some features that make them desirable alternatives to chloride pumps, which include fast recovery from inactivation and larger sized currents following activation. Arch (archaerhodopsin-3 from Halorubrum sodomense), Mac (from the fungus Leptosphaeria maculans), ArchT (an archaerhodopsin from Halorubrum strain TP009), and eBR (an enhanced version of bacteriorhodopsin from Halobacterium salinarum) are proton pumps that show robust efficiency in inhibition. Recent work has demonstrated that inhibition of eNpHR3.0-expressing neurons may render the inhibited neuron transiently more excitable due to a chloride-driven shift in the type-A γ-aminobutyric acid (GABAA) receptor reversal potential which may point towards a proton pump inhibitor as the opsin of choice for some experiments especially involving this system.
What opsin functions as a proton pump?
Which opsin, when expressed, would allow for the neuron to be excited?
- ChR2
- NpHR
- 0
- None of the above