6.4: Protein Metabolism
- Page ID
- 40962
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\dsum}{\displaystyle\sum\limits} \)
\( \newcommand{\dint}{\displaystyle\int\limits} \)
\( \newcommand{\dlim}{\displaystyle\lim\limits} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\(\newcommand{\longvect}{\overrightarrow}\)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)Section 2.2 described how proteins are synthesized. Thus, this section will focus on how proteins and amino acids are broken down. There are four protein metabolic pathways that will be covered in this section
Transamination, Deamination & Ammonia Removal as Urea
The first step in catabolizing, or breaking down, an amino acid is the removal of its amine group (-\(\ce{NH3}\)). Amine groups can be transferred or removed through transamination or deamination, respectively.
Transamination
Transamination is the transfer of an amine group from an amino acid to a keto acid (amino acid without an amine group), thus creating a new amino acid and keto acid as shown below.
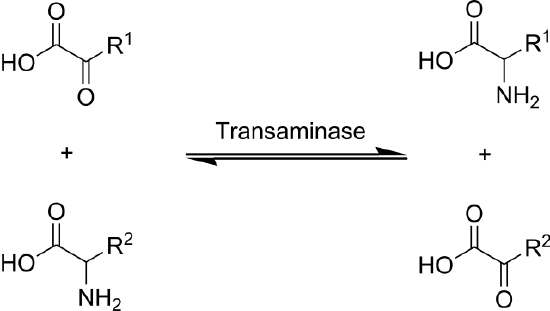
Keto acids (also known as carbon skeletons) are what remains after amino acids have had their nitrogen group removed by deamination or transamination. Transamination is used to synthesize nonessential amino acids.
Deamination
Deamination is the removal of the amine group as ammonia (\(\ce{NH3}\)), as shown below.
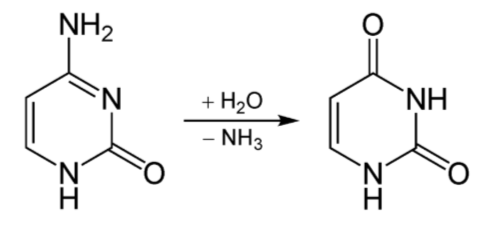
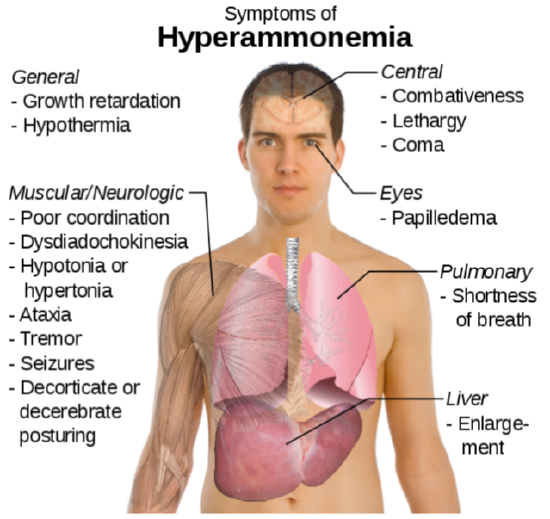
The potential problem with deamination is that too much ammonia is toxic, causing a condition known as hyperammonemia. The symptoms of this condition are shown in the following figure.
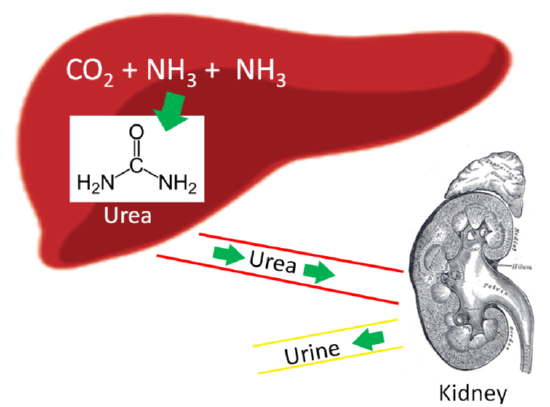
Our body has a method to safely package ammonia into a less toxic form to be excreted. This safer compound is urea, which is produced by the liver using 2 molecules of ammonia (\(\ce{NH3}\)) and 1 molecule of carbon dioxide (\(\ce{CO2}\)). Most urea is then secreted from the liver and incorporated into urine in the kidney to be excreted from the body, as shown below.
Query \(\PageIndex{1}\)
Gluconeogenesis
Gluconeogenesis is the synthesis of glucose from noncarbohydrate sources. Certain amino acids can be used for this process, which is the reason that this section is included here instead of the carbohydrate metabolism section. Gluconeogenesis is glycolysis in reverse with an oxaloacetate workaround, as shown below. Remember oxaloacetate is also an intermediate in the citric acid cycle.

Query \(\PageIndex{2}\)
Not all amino acids can be used for gluconeogenesis. The ones that can be used are termed glucogenic (red), and can be converted to either pyruvate or a citric acid cycle intermediate. Other amino acids can only be converted to either acetyl-\(\ce{CoA}\) or acetoacetyl-\(\ce{CoA}\), which cannot be used for gluconeogenesis. However, acetyl-\(\ce{CoA}\) or acetoacetyl-\(\ce{CoA}\) can be used for ketogenesis to synthesize the ketone bodies, acetone and acetoacetate. Thus, these amino acids are instead termed ketogenic (green).
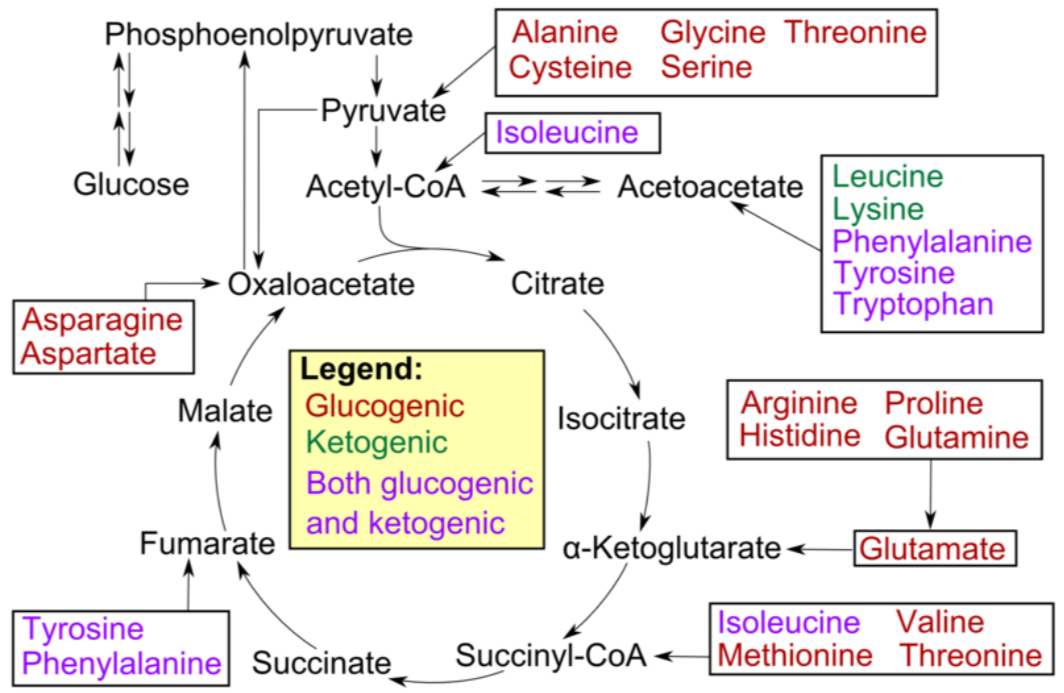
Fatty acids and ketogenic amino acids cannot be used to synthesize glucose. The transition reaction is a one-way reaction, meaning that acetyl-\(\ce{CoA}\) cannot be converted back to pyruvate. As a result, fatty acids can't be used to synthesize glucose, because beta-oxidation produces acetyl-\(\ce{CoA}\). Even if acetyl-\(\ce{CoA}\) enters the citric acid cycle, the carbons from it will eventually be completely oxidized and given off as \(\ce{CO2}\). The net result is that these carbons are not readily available to serve as keto-acids or carbon skeletons for amino acid synthesis. Some amino acids can be either glucogenic or ketogenic, depending on how they are metabolized. These amino acids are referred to as glucogenic and ketogenic (pink).
Query \(\PageIndex{3}\)
Protein Turnover/Degradation
Proteins serve a number of functions in the body, but what happens when cells, enzymes, etc. have completed their lifespan? They are recycled.
Proteins are broken down into amino acids that can be used to synthesize new proteins. There are 3 main systems of protein degradation:
- Ubiquitin-proteasome degradation
- Lysosome degradation
- Calpain degradation
Ubiquitin-Proteasome Degradation
Proteins that are damaged or abnormal are tagged with the protein ubiquitin. There are multiple protein subunits involved in the process (E1-E3), but the net result is the production of a protein (substrate) with a ubiquitin tail, as shown below.
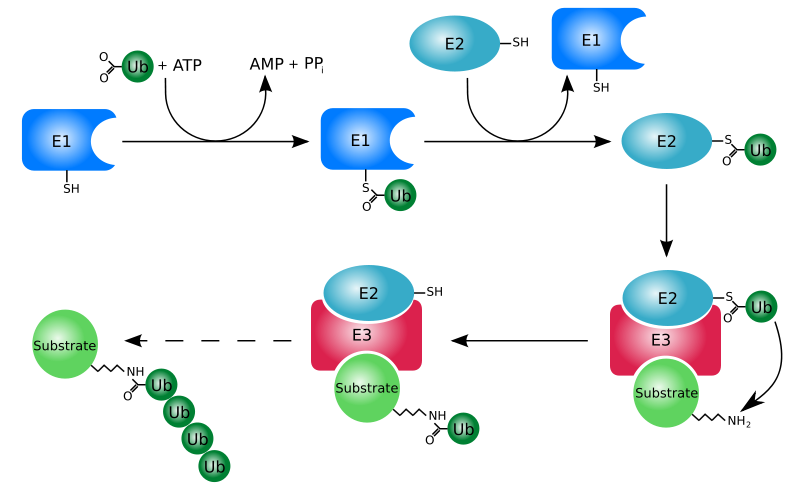
This protein then moves to the proteasome for degradation. Think of the proteasome like a garbage disposal. The ubiquitinated "trash" protein is inserted into the garbage disposal where it is broken down into its component parts (primarily amino acids). The following video illustrates this process nicely.
Lysosome Degradation
The lysosomes are organelles that are found in cells. They contain a number of proteases that degrade proteins.
Calpain Degradation
The last degradation system is the calpain system, which is not as well understood, but does require calcium.
Query \(\PageIndex{4}\)
References
- en.Wikipedia.org/wiki/File:Transaminierung.svg
- en.Wikipedia.org/wiki/File:De...ierungCtoU.png
- en.Wikipedia.org/wiki/File:Sy...rammonemia.svg
- commons.wikimedia.org/wiki/File:Liver.svg
- upload.wikimedia.org/wikipedi...ey_section.jpg
- en.Wikipedia.org/wiki/File:Urea.png
- en.Wikipedia.org/wiki/File:CellRespiration.svg
- en.Wikipedia.org/wiki/File:Am...catabolism.png
- en.Wikipedia.org/wiki/File:Ubiquitylation.svg
- en.Wikipedia.org/wiki/File:Il..._structure.jpg


