3.9: Genetic Approaches to Examine the Intact and Living Brain
- Page ID
- 10657
Labeling the Living Brain
Owing to the complexity of the mammalian brain, it has remained a major challenge to decipher the patterns of connectivity made onto and by newborn neurons as they integrate into circuits of the adult brain. With major advances in both molecular genetics and light microscopy, our ability to query not only neuronal morphologies, but also the molecular and cellular composition of individual neurons and their associated synaptic networks has become possible.
Arguably, one of the most influential contributions to contemporary neuroscience has been the use of fluorescent proteins (FPs) and their targeted expression in living neurons of the mammalian brain tissue. The wide array of FPs available provides an ever-expanding toolbox of vital reporters and gene expression tags. Applications for these proteins range from vital reporters expressed throughout the cytoplasm to subcellular protein fusion tags, which together can be used to monitor the process of circuit integration in vivo using both electrophysiological methods and fluorescent imaging.
Beyond merely marking cells for identification, a number of other methods have been developed to exploit the vital properties of FPs to investigate neuronal properties. For example, superecliptic pHluorin, which fluoresces at neutral pH but is quenched at acidic pH, can be used to monitor the trafficking and exchange of intracellular compartments within neurons. This variant allows direct imaging of membrane dynamics, exocytosis and endocytosis of synaptic receptors, and neurotransmitter release in vitro and in vivo. More recently, a new method termed GFP reconstitution across synaptic partners (GRASP) shows promise for revealing synaptic interactions between contacting neurons. By tethering split GFP fragments to separate pre- and post-synaptic proteins, reconstitution of GFP fluorescence can be observed when genetically targeted cells form synaptic pairs. Although this technology has been successfully applied to reveal invertebrate synapses, it has yet to be demonstrated in rodents (Gordon and Scott, 2009).
The range of FP reporters for visualizing neuronal morphologies, cellular dynamics, and synapse function continues to expand. However, perhaps the single-most limiting factor for using FPs in neuroscience is our incomplete knowledge of neuronal gene regulation. Often transgenic reporters fail to recapitulate endogenous patterns of gene expression, or such patterns are too broad to identify neuronal subtypes with cellular precision.
Trans-Synaptic Circuit Tracing
A major goal toward understanding mechanisms of neuronal development, synapse formation, and circuit wiring has been to elucidate nodes and patterns of synaptic connectivity. A creative angle to address this challenge has been the incorporation of genes encoding FPs and FP-fusion proteins into neurotropic viral vectors, which show the innate ability to infect neurons and trans-synaptically spread throughout the nervous system (Kuypers and Ugolini, 1990; Callaway, 2008).
Two types of viruses that have been broadly employed for this purpose include rabies and herpes. Herpes viruses belong to a family of double-stranded DNA viruses, while rabies belongs to a family of negative-strand RNA viruses (Voyles, 1993). Although evolutionarily different, they are both endowed with the unique ability to bind to and infect neuronal cells. This cell type-specific infectivity is conferred to the viruses via their mature enveloped coat particles, which are made of both host membrane and virally encoded glycoproteins. The composite envelope proteins are the determinants that mediate neuronal membrane recognition and subsequent neuron-to-neuron infection by binding to membrane surface receptors.
Herpes viruses have been used to label neural circuits for years. Two common tracing strains are herpes simplex virus-1 (HSV-1; Lilley et al., 2001) and pseudorabies virus (PRV; Enquist, 2002). Both of these variants predominantly spread in a retrograde direction, and each has been effectively applied to dissect synapse and circuit connections in the rodent brain (Callaway, 2008). However, one limitation to using the herpes viruses for circuit analysis is polysynaptic spreading. Due to the vast cohort of cell types within brain tissue, the number of synapses formed on each of those cells, and the high degree of interconnectivity in intact neural circuits, this approach still poses a challenge to dissect precise patterns of neural connectivity. To simplify trans-synaptic circuit analysis, Wickersham et al. (2007b) devised a clever coat protein complementation strategy that allows for monosynaptic tracing of neuronal connections using a pseudotyped rabies virus (RV). Not to be confused with PRV (which as stated above is actually a herpes virus), pseudotyping a viral particle refers to synthetically modifying the viral envelope to recognize a foreign receptor not normally present on the membranes of mammalian neurons. The strategy will be briefly discussed below, and for further reference also see Wickersham et al. (2007a), Arenkiel and Ehlers (2009), Hasenstaub and Callaway (2010).
The RV gene encoding its glycoprotein (termed G) has been the primary target for genetic modification and RV vector engineering. Removal of G from the RV genome renders the virus both incapable of generating infective particles and replication incompetent. However, even in the absence of the native glycoprotein gene, RV is still capable of expressing its genome. Thus, G can be replaced with sequences encoding FPs or FP-tagged biomolecules to generate RV vectors for vital reporter expression (Wickersham et al., 2007a). To make these replication incompetent viruses useful for circuit tracing studies, they must be “armed” by providing an envelope in trans by propagating and packaging the particles in vitrousing cell lines engineered to synthesize the required glycoprotein.
To perform monosynaptic circuit tracing and target FP-expressing RV to desired neuronal subsets, the particles can first be pseudotyped with the foreign coat protein EnvA from avian sarcoma leukosis virus, which specifically binds to a class of avian membrane proteins called TVA receptors (Barnard et al., 2006). Genetic targeting of neuronal subsets for TVA expression directs RV infection to only those neurons. To facilitate monosynaptic tracing, Wickersham et al. (2007a)added a clever twist on this approach. By introducing a plasmid that encodes the wildtype RV G-protein, the disarmed EGFP-expressing virus is now able to undergo one round of subsequent infection to presynaptic partners of TVA-targeted neurons. Since only the initially infected neuron contains G, viral spread ceases after one round of monosynaptic jumping. Including a plasmid encoding a red FP allows the cell originally targeted for infection to be identified amongst the monosynaptic network of GFP labeled cells (Figure \(\PageIndex{1}\)). Of course it must be considered that true monosynaptic tracing is dependent on targeting individual neurons for the expression of G. If for example synaptically coupled cells both harbor G, but only one of them serves as the primary source cell of TVA-mediated infection, then viral spread can become multisynaptic through subsequent rounds of viral packaging in presynaptic partners. Monosynaptic tracing control thus depends directly upon the precision of neuronal targeting for the RV tracing components.
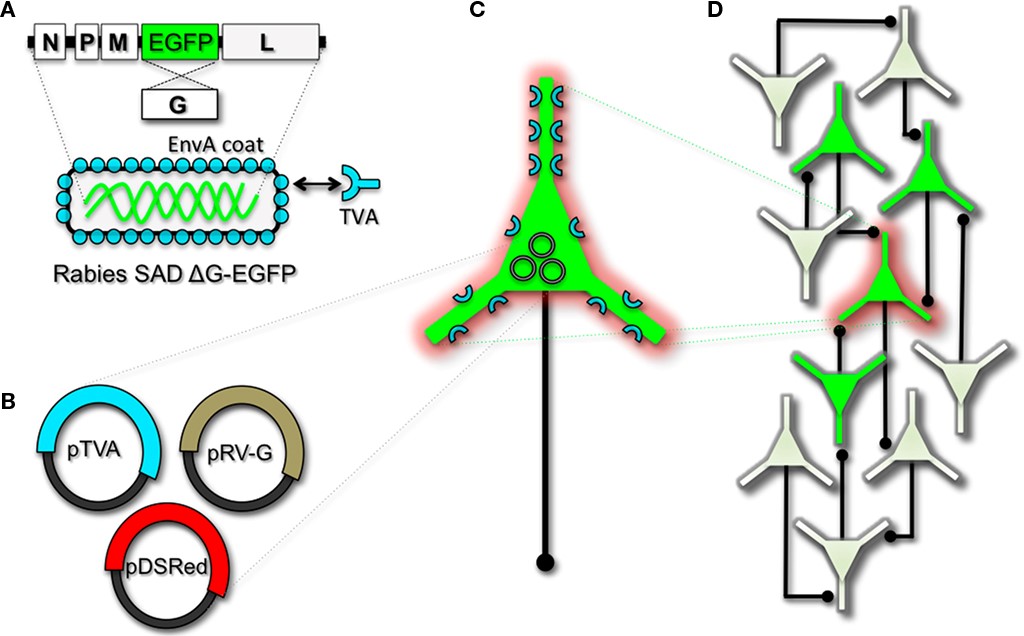
Figure \(\PageIndex{1}\). Engineering and pseudotyping rabies virus (RV) for transneuronal tracing. (A) RV can be genetically engineered to express EGFP by replacing the genomic sequence encoding the G coat protein. The genetically modified G-deletion mutant RV must be propagated in vitro to supply a coat protein. The particle can thus be pseudotyped by providing a foreign coat protein such as EnvA, which originates from the avian leukosis virus and binds specifically to it its cognate receptor TVA. EnvA pseudotyped RV can be used to selectively infect neurons that have been genetically targeted for TVA expression. By including additional constructs that encode the wildtype G-capsid protein and a red-colored “cell fill” (B), the modified RV can be genetically targeted to individual neurons for restricted circuit mapping and monosynaptic tracing (C). Since no endogenous receptors exist in the mammalian brain for EnvA, only neurons that are programmed to express TVA are capable of being infected by the EnvA pseudotyped virions. Because the wildtype G-protein sequence has been deleted from the RV genome, G must be supplied by complementation to allow trans-synaptic spread from the neurons targeted for infection. (D) Viral spread ceases monosynaptically due to the absence of G in unmodified neuronal populations.
This new technology now makes it feasible to dissect complicated patterns of neuronal connectivity with synaptic precision (Stepien et al., 2010; Weible et al., 2010). Targeting adult-born neurons for monosynaptic circuit tracing holds certain promise toward elucidating the numbers, types, and synaptic inputs that might usher and/or promote the formation and maintenance of functional circuit integration. Unfortunately however, much needs to be learned about the viral mechanisms of infectivity, trans-synaptic propagation, and replication to make viral tracing methods broadly applicable for detailed circuit analysis throughout the nervous system. For example, one major limitation to viral-mediated circuit tracing using either HSV or RV type vectors is the inevitable deterioration of neuronal cell health with time (Callaway, 2008). Whereas the HSV particles show rapid and high levels of expression within 1–2 days, they also show a lytic-type phase of replication that induces neuronal loss within 1–2 weeks. Although most neurons appear to tolerate RV infection for longer periods of time, they too eventually show signs of dysfunction and poor health beyond 2 weeks. In addition, not much is known regarding the exact tropism for the various viruses to infect particular subtypes of neurons. Although it is clear that viral particles can cross axo-dendritic, dendro-dendritic, glutamatergic, and GABAergic synapses (Willhite et al., 2006; Wickersham et al., 2007b; Stepien et al., 2010; Miyamichi et al., 2011; Rancz et al., 2011), the different efficacies of transfer have not been determined. Preferential binding of viral particles to different types of presynaptic proteins must exist, which would ultimately result in more efficient transfer of viruses between certain synaptic pairs. This information is currently unknown, thus it remains a challenge to reliably perform unbiased quantitative circuit analysis using viruses over extended periods of time.
Although current trans-synaptic circuit tracing methods are in their infancy, with further understanding of the viral mechanisms, and a subsequent “re-tooling” of existing vectors, one can easily imagine that this experimental avenue for intact circuit mapping will become indispensable. Moreover, this methodology holds definite promise to address outstanding questions in adult neurogenesis, ranging from identification of the types of connections that are dynamically made and broken during circuit development, to exposing the complete cohort of input types that are observed in mature circuits within the intact brain.
Manipulating Cell and Circuit Activity
Earmarking neuronal subsets and their associated networks for has been invaluable toward our current understanding of neuronal morphologies and circuit architecture. However, to fully understand the cellular and molecular mechanisms that guide adult-born neuron synapse formation and circuit integration, we must be able to probe neuronal connectivity. Recent advances in genetically encoded actuators now provide this possibility. Technologies such as heterologous receptor or channel expression, optogenetics, and genetically encoded synaptic toxins are beginning to allow functional circuit mapping with synaptic precision (Luo et al., 2008; Arenkiel and Ehlers, 2009; Figure \(\PageIndex{3}\)). By targeting pre- or post-synaptic cell types for activity manipulations, coupled with functional imaging and/or electrophysiological recordings, it is now possible to genetically dissect circuit nodes by monitoring evoked synthetic output responses. Some of the earliest efforts to genetically control neuronal output relied on engineered expression of heterologous receptors in neurons that normally do not show their presence. For example, expression of modified opiate receptors in the brains of transgenic mice showed that introducing synthetic exogenous ligands could activate neuronal subsets (Zhao et al., 2003). To date, numerous variations on this theme have proven effective for both driving neuronal excitability and inhibition. Complementary strategies to these methods have been to genetically express small-molecules for inactivation of synaptic transmission (Karpova et al., 2005), or toxins that disrupt synaptic transmission (Harms et al., 2005; Ehlers et al., 2007).
Genetic strategies to mark and manipulate neurons and circuits
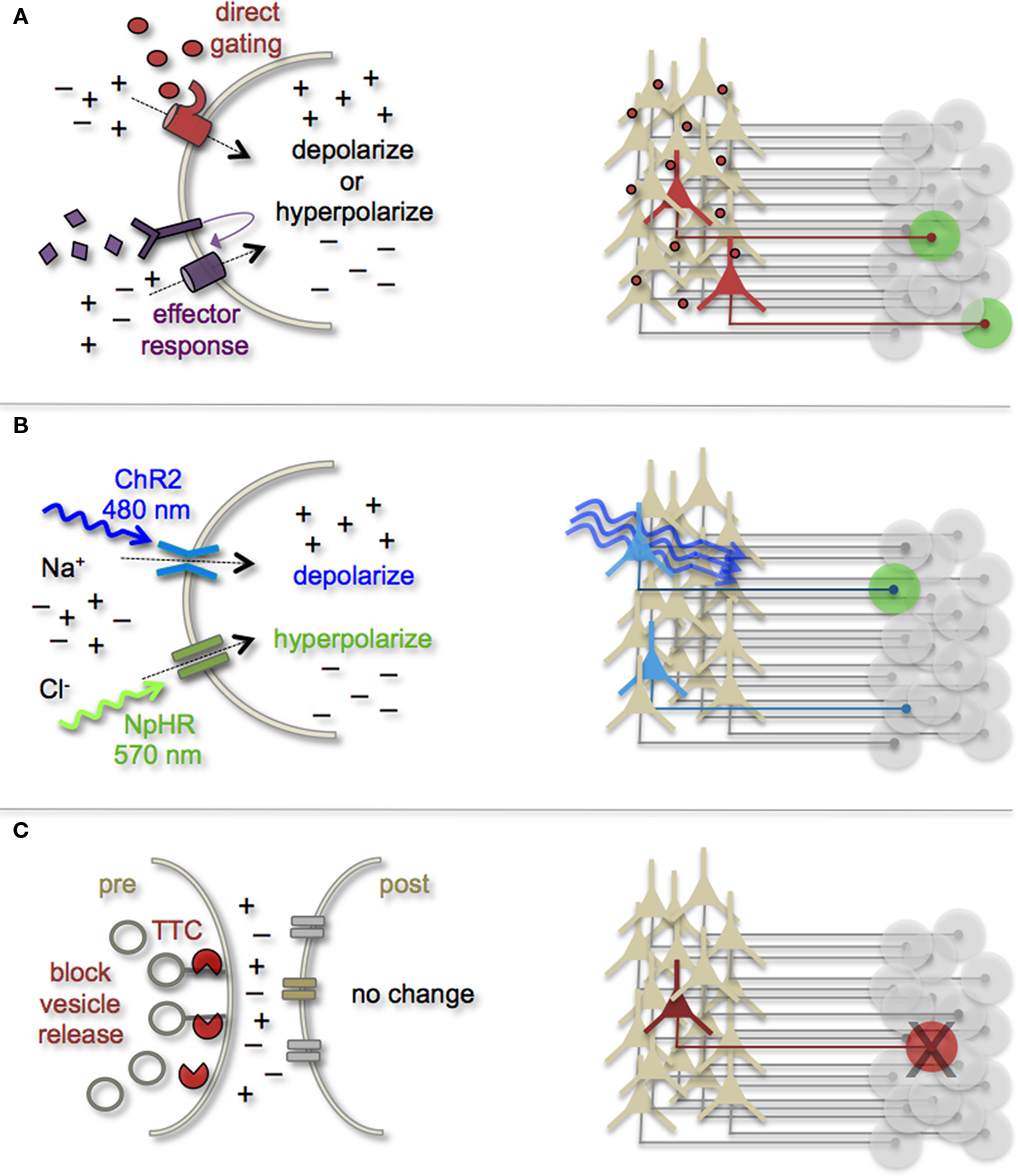
Figure \(\PageIndex{3}\). (A) Neurons can be targeted for heterologous receptor expression. These foreign receptors can be either directly or indirectly gated by application of exogenous ligands (depicted as red ovals and purple diamonds). Left: heterologous receptor activation via application of synthetic ligands can be used to change a neuron’s ionic equilibrium and thus firing properties. Right: depending on molecular properties, exogenous ligands spread variably throughout brain tissue. All neurons expressing the heterologous receptors are capable of being activated and driving target cell responses (represented as green circles). (B) Expression of light-gated channels can be used to modulate neuronal firing with photons. Left: ChR-2 is a non-selective cation channel that responds optimally to blue light. Photostimulating this channel results in positive inward currents, depolarization, and neuronal firing. NpHR is a photoactive chloride pump that responds optimally to greenish-yellow light. Photostimulating this pump protein results in negative inward currents, hyperpolarization, and neuronal silencing. Right: FP-fusion reporters can be used to identify cells that express photoresponsive proteins (represented by blue coloring). Only neurons expressing the photoresponsive channels and receive photons show light activated modulation, whereas downstream circuit targets can be monitored for post-synaptic photo responses (green coloring). (C) Targeted expression of synaptic toxins in neurons can be used to block synaptic vesicle release and inhibit neurotransmission to post-synaptic targets. TTC, tetanus toxin.