10.4.1E: Clonal Selection and Clonal Expansion
- Page ID
- 77753
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\( \newcommand{\id}{\mathrm{id}}\) \( \newcommand{\Span}{\mathrm{span}}\)
( \newcommand{\kernel}{\mathrm{null}\,}\) \( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\) \( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\) \( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\id}{\mathrm{id}}\)
\( \newcommand{\Span}{\mathrm{span}}\)
\( \newcommand{\kernel}{\mathrm{null}\,}\)
\( \newcommand{\range}{\mathrm{range}\,}\)
\( \newcommand{\RealPart}{\mathrm{Re}}\)
\( \newcommand{\ImaginaryPart}{\mathrm{Im}}\)
\( \newcommand{\Argument}{\mathrm{Arg}}\)
\( \newcommand{\norm}[1]{\| #1 \|}\)
\( \newcommand{\inner}[2]{\langle #1, #2 \rangle}\)
\( \newcommand{\Span}{\mathrm{span}}\) \( \newcommand{\AA}{\unicode[.8,0]{x212B}}\)
\( \newcommand{\vectorA}[1]{\vec{#1}} % arrow\)
\( \newcommand{\vectorAt}[1]{\vec{\text{#1}}} % arrow\)
\( \newcommand{\vectorB}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vectorC}[1]{\textbf{#1}} \)
\( \newcommand{\vectorD}[1]{\overrightarrow{#1}} \)
\( \newcommand{\vectorDt}[1]{\overrightarrow{\text{#1}}} \)
\( \newcommand{\vectE}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash{\mathbf {#1}}}} \)
\( \newcommand{\vecs}[1]{\overset { \scriptstyle \rightharpoonup} {\mathbf{#1}} } \)
\( \newcommand{\vecd}[1]{\overset{-\!-\!\rightharpoonup}{\vphantom{a}\smash {#1}}} \)
\(\newcommand{\avec}{\mathbf a}\) \(\newcommand{\bvec}{\mathbf b}\) \(\newcommand{\cvec}{\mathbf c}\) \(\newcommand{\dvec}{\mathbf d}\) \(\newcommand{\dtil}{\widetilde{\mathbf d}}\) \(\newcommand{\evec}{\mathbf e}\) \(\newcommand{\fvec}{\mathbf f}\) \(\newcommand{\nvec}{\mathbf n}\) \(\newcommand{\pvec}{\mathbf p}\) \(\newcommand{\qvec}{\mathbf q}\) \(\newcommand{\svec}{\mathbf s}\) \(\newcommand{\tvec}{\mathbf t}\) \(\newcommand{\uvec}{\mathbf u}\) \(\newcommand{\vvec}{\mathbf v}\) \(\newcommand{\wvec}{\mathbf w}\) \(\newcommand{\xvec}{\mathbf x}\) \(\newcommand{\yvec}{\mathbf y}\) \(\newcommand{\zvec}{\mathbf z}\) \(\newcommand{\rvec}{\mathbf r}\) \(\newcommand{\mvec}{\mathbf m}\) \(\newcommand{\zerovec}{\mathbf 0}\) \(\newcommand{\onevec}{\mathbf 1}\) \(\newcommand{\real}{\mathbb R}\) \(\newcommand{\twovec}[2]{\left[\begin{array}{r}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\ctwovec}[2]{\left[\begin{array}{c}#1 \\ #2 \end{array}\right]}\) \(\newcommand{\threevec}[3]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\cthreevec}[3]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \end{array}\right]}\) \(\newcommand{\fourvec}[4]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\cfourvec}[4]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \end{array}\right]}\) \(\newcommand{\fivevec}[5]{\left[\begin{array}{r}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\cfivevec}[5]{\left[\begin{array}{c}#1 \\ #2 \\ #3 \\ #4 \\ #5 \\ \end{array}\right]}\) \(\newcommand{\mattwo}[4]{\left[\begin{array}{rr}#1 \amp #2 \\ #3 \amp #4 \\ \end{array}\right]}\) \(\newcommand{\laspan}[1]{\text{Span}\{#1\}}\) \(\newcommand{\bcal}{\cal B}\) \(\newcommand{\ccal}{\cal C}\) \(\newcommand{\scal}{\cal S}\) \(\newcommand{\wcal}{\cal W}\) \(\newcommand{\ecal}{\cal E}\) \(\newcommand{\coords}[2]{\left\{#1\right\}_{#2}}\) \(\newcommand{\gray}[1]{\color{gray}{#1}}\) \(\newcommand{\lgray}[1]{\color{lightgray}{#1}}\) \(\newcommand{\rank}{\operatorname{rank}}\) \(\newcommand{\row}{\text{Row}}\) \(\newcommand{\col}{\text{Col}}\) \(\renewcommand{\row}{\text{Row}}\) \(\newcommand{\nul}{\text{Nul}}\) \(\newcommand{\var}{\text{Var}}\) \(\newcommand{\corr}{\text{corr}}\) \(\newcommand{\len}[1]{\left|#1\right|}\) \(\newcommand{\bbar}{\overline{\bvec}}\) \(\newcommand{\bhat}{\widehat{\bvec}}\) \(\newcommand{\bperp}{\bvec^\perp}\) \(\newcommand{\xhat}{\widehat{\xvec}}\) \(\newcommand{\vhat}{\widehat{\vvec}}\) \(\newcommand{\uhat}{\widehat{\uvec}}\) \(\newcommand{\what}{\widehat{\wvec}}\) \(\newcommand{\Sighat}{\widehat{\Sigma}}\) \(\newcommand{\lt}{<}\) \(\newcommand{\gt}{>}\) \(\newcommand{\amp}{&}\) \(\definecolor{fillinmathshade}{gray}{0.9}\)- Briefly describe the process of clonal selection and clonal expansion.
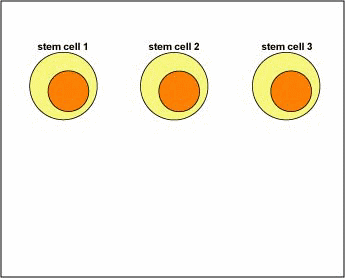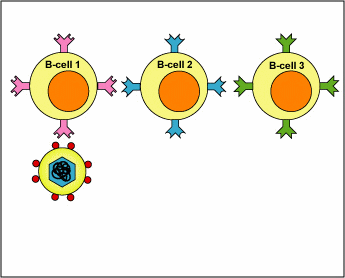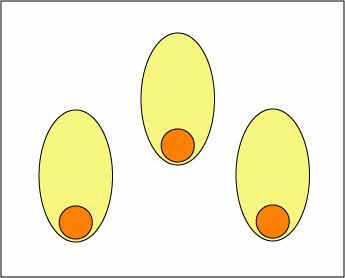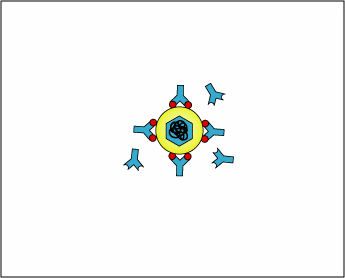
As mentioned above, during early differentiation of naive B-lymphocytes in the bone marrow, each B-lymphocyte becomes genetically programmed to make an antibody with a unique antigen-binding site (Fab) through a series of gene translocations, and molecules of that antibody are put on its surface to function as the B-cell receptor (Figure \(\PageIndex{1}\)).
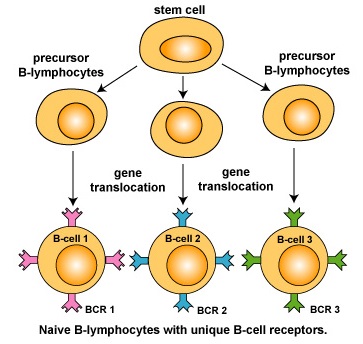 Figure \(\PageIndex{1}\): Clonal Selection, Step-1. During its development, each B-lymphocyte becomes genetically programmed, through a process called gene translocation, to make a unique B-cell receptor. Molecules of that B-cell receptor are placed on its surface where it can react with epitopes of an antigen.
Figure \(\PageIndex{1}\): Clonal Selection, Step-1. During its development, each B-lymphocyte becomes genetically programmed, through a process called gene translocation, to make a unique B-cell receptor. Molecules of that B-cell receptor are placed on its surface where it can react with epitopes of an antigen.
When an antigen encounters the immune system, its epitopes eventually will react only with B-lymphocytes with B-cell receptors on their surface that more or less fit and this activates those B-lymphocytes. This process is known as clonal selection (Figure \(\PageIndex{2}\)).
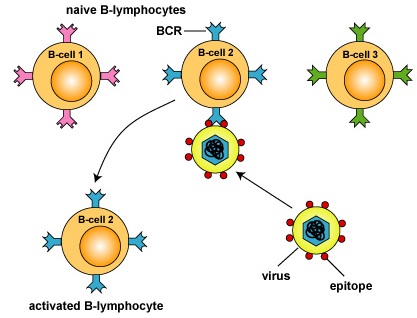 Figure \(\PageIndex{2}\): Clonal Selection, Step-2. A B-lymphocyte with an appropriately fitting B-cell receptor can now react with epitopes of an antigen having a corresponding shape. This activates the B-lymphocyte.
Figure \(\PageIndex{2}\): Clonal Selection, Step-2. A B-lymphocyte with an appropriately fitting B-cell receptor can now react with epitopes of an antigen having a corresponding shape. This activates the B-lymphocyte.
Cytokines produced by effector T4-helper lymphocytes enable those activated B-lymphocytes to rapidly proliferate to produce large clones of thousands of identical B-lymphocytes. In this way, even though only a few B-lymphocytes in the body may have an antibody molecule able to fit a particular epitope, eventually many thousands of cells are produced with the right specificity. This is referred to as clonal expansion (Figure \(\PageIndex{3}\)).
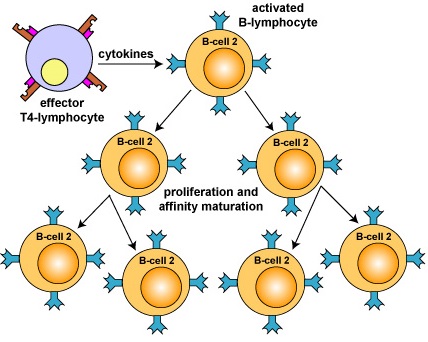 Figure \(\PageIndex{3}\): Clonal Expansion. Cytokines from an effector T4-lymphocyte now enable the activated B-lymphocyte to proliferate into a large clone of identical B-lymphocytes. During this time, "fine-tuning" of the B-cell receptor occurs through affinity maturation.
Figure \(\PageIndex{3}\): Clonal Expansion. Cytokines from an effector T4-lymphocyte now enable the activated B-lymphocyte to proliferate into a large clone of identical B-lymphocytes. During this time, "fine-tuning" of the B-cell receptor occurs through affinity maturation.
Furthermore, as the B-lymphocytes proliferate, they undergo affinity maturation as a result of somatic hypermutations. This allows the B-lymphocytes to "fine-tune" the shape of the antibody for better fit with the original epitope. B-lymphocytes having better fitting B-cell receptor on their surface bind epitope longer and more tightly allowing these cells to selectively replicate. Eventually these variant B-lymphocytes differentiate intoplasma cells that synthesize and secrete vast quantities of antibodies that have Fab sites which fit the original epitope very precisely (Figure \(\PageIndex{4}\)). It generally takes 4-5 days for a naive B- lymphocyte that has been activated to complete clonal expansion and differentiate into effector B-lymphocytes.
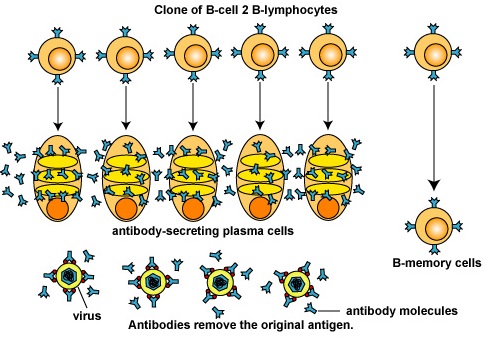 Figure \(\PageIndex{4}\): Differentiation of B-lymphocytes into Plasma Cells and B-Memory Cells. The B-lymphocytes now differentiate into antibody-secreting B-lymphocytes and plasma cells that secrete large quantities of antibodies that "fit" the original epitope. Some B-lymphocytes differentiate into B-memory cells capable of anamnestic response.
Figure \(\PageIndex{4}\): Differentiation of B-lymphocytes into Plasma Cells and B-Memory Cells. The B-lymphocytes now differentiate into antibody-secreting B-lymphocytes and plasma cells that secrete large quantities of antibodies that "fit" the original epitope. Some B-lymphocytes differentiate into B-memory cells capable of anamnestic response.
A single activated B-lymphocyte can, within seven days, give rise to approximately 4000 antibody-secreting cells. Over 2000 antibody molecules can be produced per plasma cell per second for typically up to four to five days. The B-memory cells that eventually form also have these high affinity antibodies on their surface.
During its development, each B-lymphocyte becomes genetically programmed, through a process called gene translocation, to make a unique antibody molecule that will function as a B-cell receptor. Molecules of that antibody are then placed on the cell's surface where it can react with epitopes of an antigen. A B-lymphocyte with an appropriately fitting B-cell receptor can now react with epitopes of an antigen having a corresponding shape. This activates the B-lymphocyte. Cytokines from an activated T4-lymphocyte now enable the activated B-lymphocyte to proliferate into a large clone of identical B-lymphocytes. During this time, "fine-tuning" of the B-cell receptor occurs as a result of affinity maturation. The B-lymphocytes now differentiate into antibody-secreting B-lymphocytes and plasma cells that secrete large quantities of antibodies "fitting" the original epitope. Some B-lymphocytes differentiate into B-memory cells capable of anamnestic response.
As with naive B-lymphocytes, during its development, each naive T4-lymphocyte becomes genetically programmed by gene-splicing reactions similar to those in B-lymphocytes, to produce a TCR with a unique specificity. Identical molecules of that TCR are placed on its surface where they are able to bind an epitope/MHC-II complex on an antigen-presenting dendritic cell with a corresponding shape (Figure \(\PageIndex{5}\)). This is clonal selection of the T4-lymphocytes that are required for the body's response to T-dependent antigens.
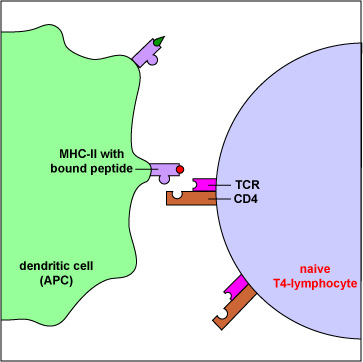 Figure \(\PageIndex{5}\): A Naive T4-Lymphocyte Recognizing Epitope/MHC-II on an Antigen-Presenting Dendritic Cell. Exogenous antigens are those from outside cells of the body. Examples include bacteria, free viruses, yeasts, protozoa, and toxins. These exogenous antigens enter antigen-presenting dendriticcells through phagocytosis. The microbes are engulfed and placed in a phagosome. After lysosomes fuse with the phagosome, protein antigens are degraded by proteases into a series of peptides. These peptides eventually bind to grooves in MHC-II milecules and are transported to the surface of the dendritic cell. Naive T4-lymphocytes are then able to recognize peptide/MHC-II complexes by means of their T-cell receptors (TCRs) and CD4 molecules.
Figure \(\PageIndex{5}\): A Naive T4-Lymphocyte Recognizing Epitope/MHC-II on an Antigen-Presenting Dendritic Cell. Exogenous antigens are those from outside cells of the body. Examples include bacteria, free viruses, yeasts, protozoa, and toxins. These exogenous antigens enter antigen-presenting dendriticcells through phagocytosis. The microbes are engulfed and placed in a phagosome. After lysosomes fuse with the phagosome, protein antigens are degraded by proteases into a series of peptides. These peptides eventually bind to grooves in MHC-II milecules and are transported to the surface of the dendritic cell. Naive T4-lymphocytes are then able to recognize peptide/MHC-II complexes by means of their T-cell receptors (TCRs) and CD4 molecules.
In response to cytokines, these activated T4-lymphocytes now rapidly proliferate and differentiate into effector T4-lymphocytes. This is clonal expansion of the T4-lymphocytes. Before an antigen enters the body, the number of naive T4-lymphocytes specific for any particular antigen is between 1 in 105 to 106 lymphocytes. After antigen exposure, the number of T4-lymphocytes specific for that antigen may increase to 1 in 100 to 1000 lymphocytes.
Summary
- Each naïve B-cell becomes genetically programmed to make an antibody with a unique antigen-binding site (Fab) through a series of gene translocations, and molecules of that antibody are put on its surface to function as the B-cell receptor.
- When an antigen encounters the immune system, its epitopes eventually will react only with B-lymphocytes with B-cell receptors on their surface that more or less fit and this activates those B-lymphocytes. This process is known as clonal selection.
- Cytokines produced by activated T4-helper lymphocytes enable those activated B-lymphocytes to rapidly proliferate to produce large clones of thousands of identical B-lymphocytes.
- In this way, even though only a few B-lymphocytes in the body may have an antibody molecule able to fit a particular epitope, eventually eventually many thousands of cells are produced with the right specificity. This is referred to as clonal expansion.
Questions
Study the material in this section and then write out the answers to these questions. Do not just click on the answers and write them out. This will not test your understanding of this tutorial.
- Briefly describe the process of clonal selection and clonal expansion. (ans)
- Multiple Choice (ans)



